Repertoire Sequencing
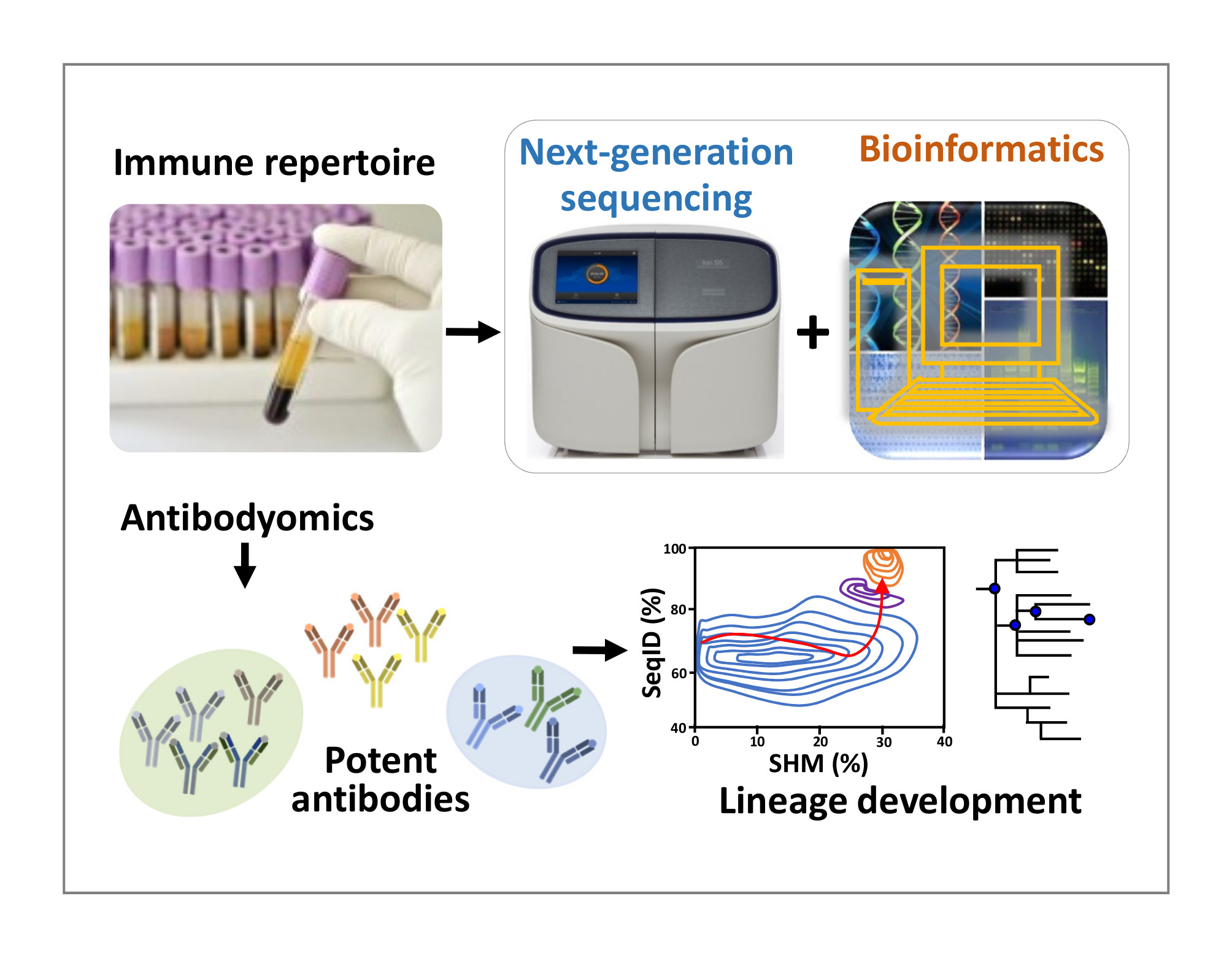
A series of novel NGS technologies have been developed for B cell repertoire analysis. Specifically, 5?-RACE PCR, reverse primers targeting constant regions, and 600bp long-read NGS are combined to capture B cell repertoires in an unbiased manner, which is key to repertoire profiling. A single-molecule barcoding strategy has been devised to reduce amplification noise and sequencing error. NGS data can be processed by the Antibodyomics pipeline to identify functional antibodies and dissect antibody lineages. These repertoire technologies are applicable to humans, macaques, rabbits, and mice.
Digital Panning
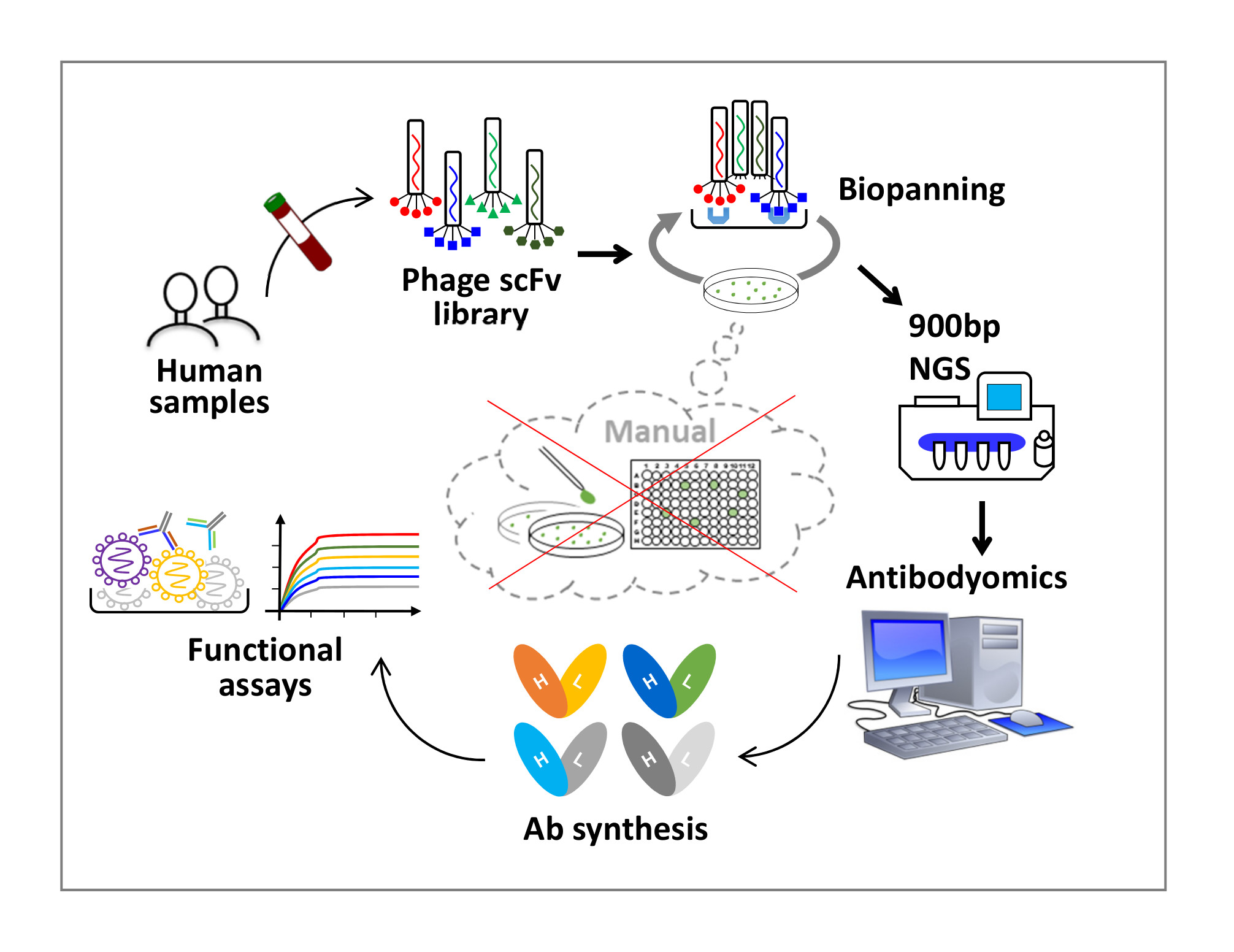
Phage display has been widely used to produce monoclonal antibodies (mAbs). Single-chain variable fragments (scFv) can be displayed on the phage surface and subjected to a selection process known as biopanning. By combining phage display of scFv libraries, ultra-long-read NGS (900bp) , and H/L-paired Antibodyomics, we developed the “digital panning” technology for antibody discovery. By nature, this technology is both analytical and deterministic, as it can capture full-length (>800bp) scFvs at each step of the panning process and select representative scFv clones based on their frequency in the library.
Epitope Scaffolding
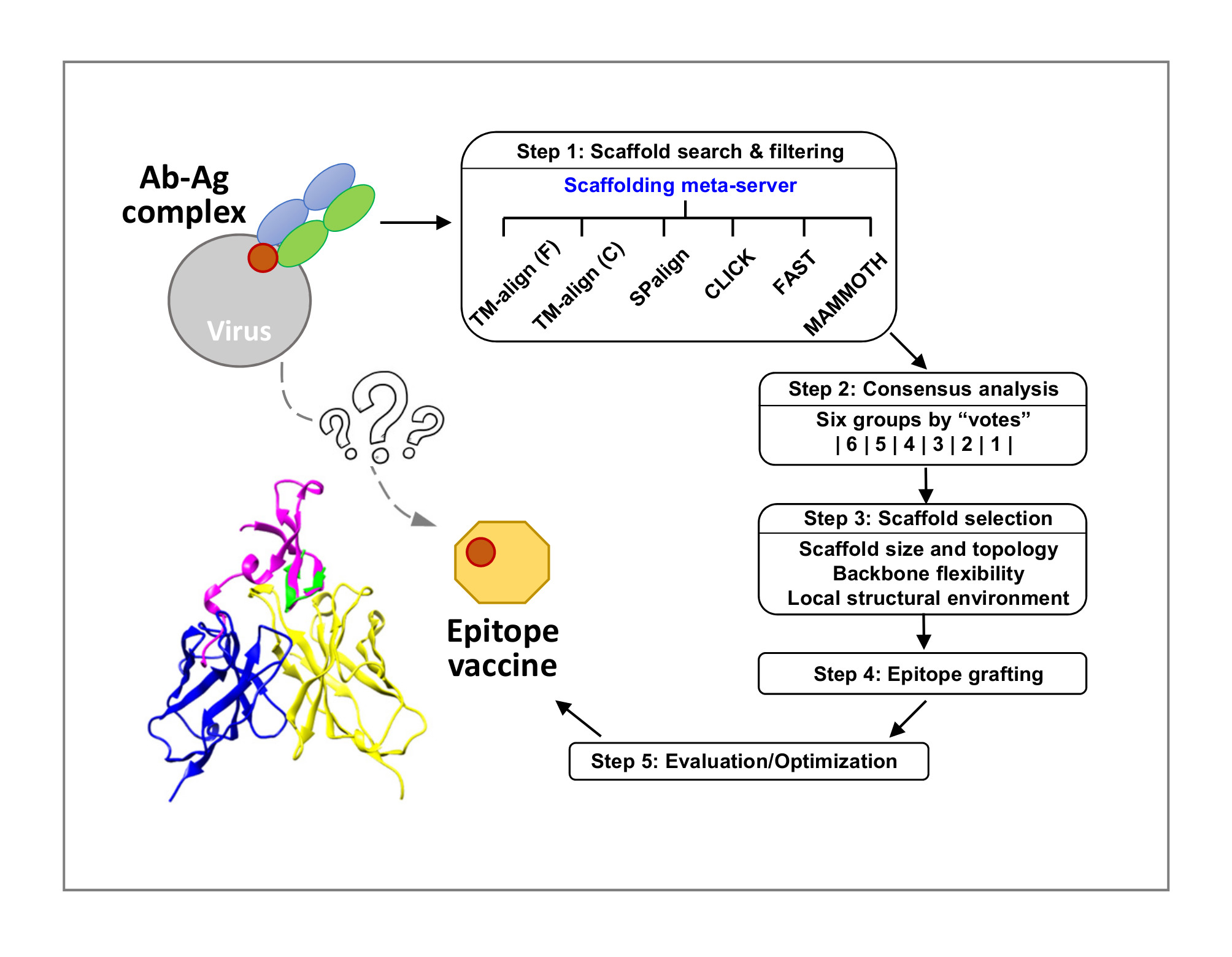
In vaccine design, a conserved neutralizing epitope on the virus surface protein can be “grafted” onto heterologous protein scaffolds to focus B cell response to the epitope target. To design such “epitope-focused” antigens, a scaffolding meta-server has been developed that combines six structural alignment (SA) algorithms to search for compatible scaffolds in the Protein Database Bank (PDB), and utilizes a consensus-based voting scheme to prioritize scaffolds for further selection and optimization. This method has been applied to several epitopes on HIV and HCV surface proteins.
UFO Trimer Design
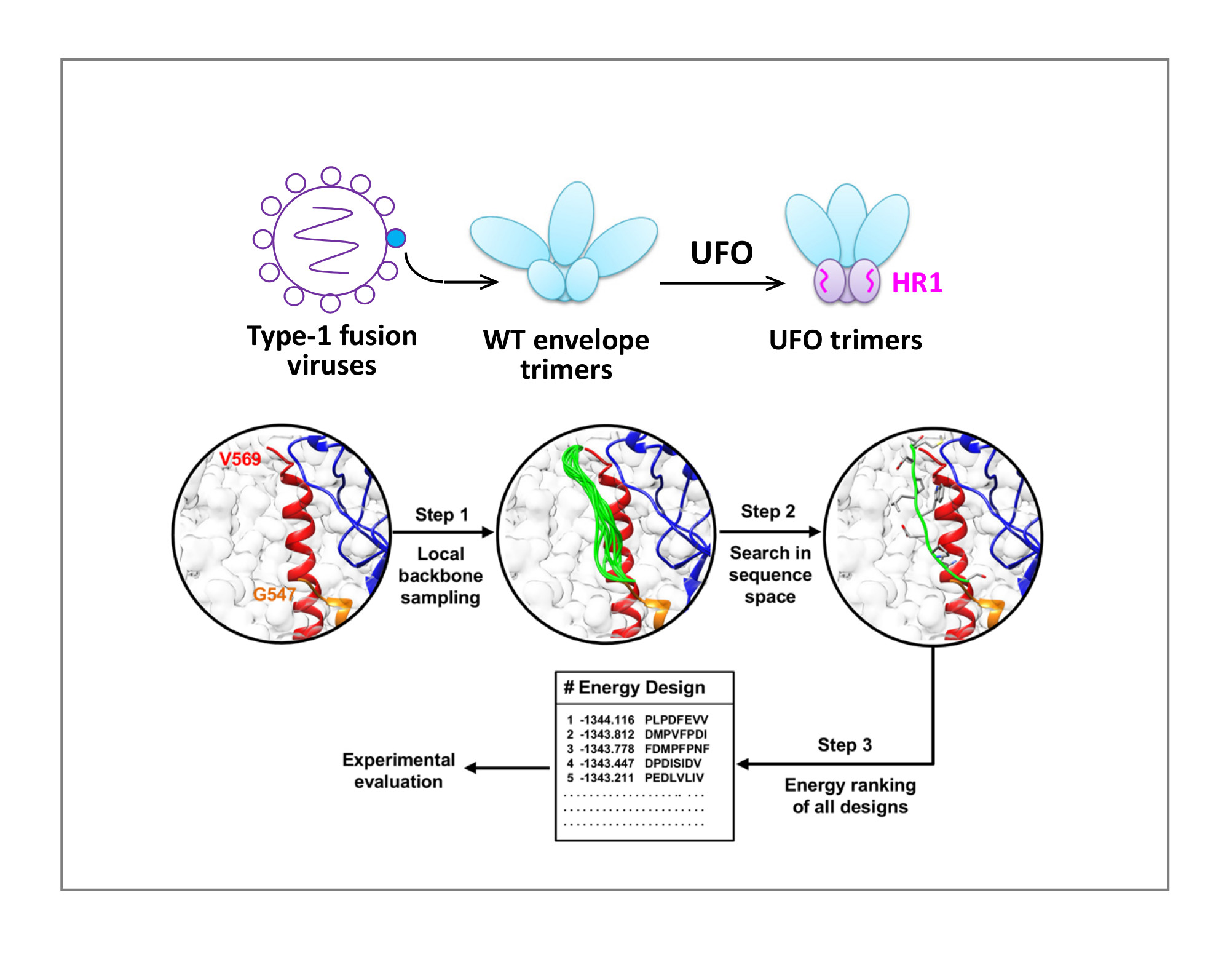
Uncleaved, prefusion-optimized (UFO) design can be used to stabilize trimeric envelope (Env) glycoproteins of diverse viruses utilizing the class-I fusion mechanism. The UFO platform was originally developed and validated in the context of HIV trimer design based on the hypothesis that an N-terminal region of heptad repeat 1 (HR1, aa547-569) in gp41 is the primary cause of HIV Env metastability. Computational redesign of this HR1 region with an ensemble-based protein design method produced UFO trimers with high yield, purity, and stability, as well as native-like Env conformation and antigenicity.
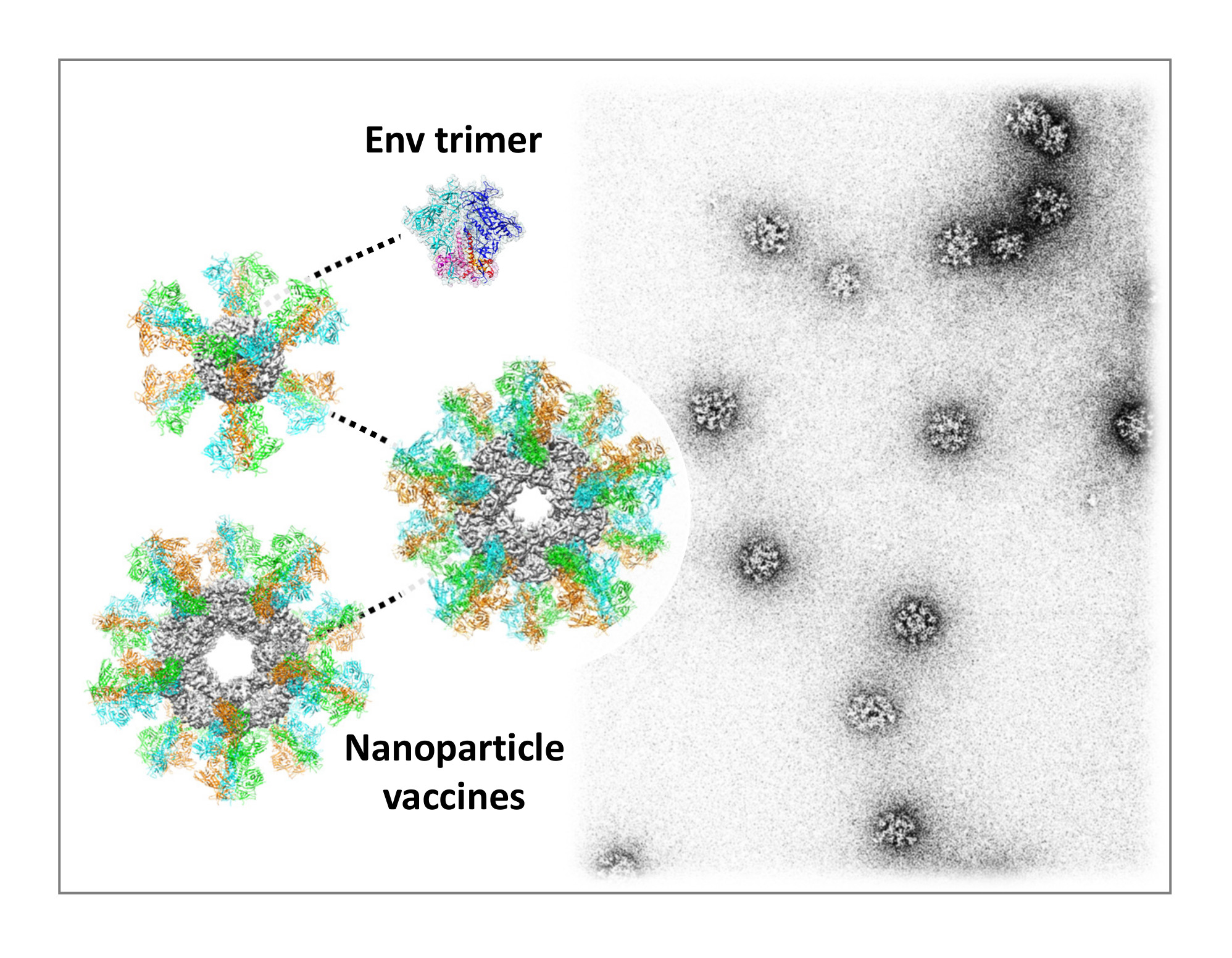
Nanoparticle Display
Single subunit vaccines are often less effective than virus-like particle (VLP) vaccines, which can induce potent and long-lasting immune responses due to their large size and dense display of surface antigens. Self-assembling nanoparticles with VLP-like molecular traits thus provide robust platforms for vaccine development without the need for complicated purification process typical of VLPs. Three nanoparticle platforms, 24-meric ferritin (13nm) and 60-meric E2p and NC1 (23-25nm), have been optimized to present stabilized HIV, HCV, Flu, and Ebola antigens as vaccine candidates.
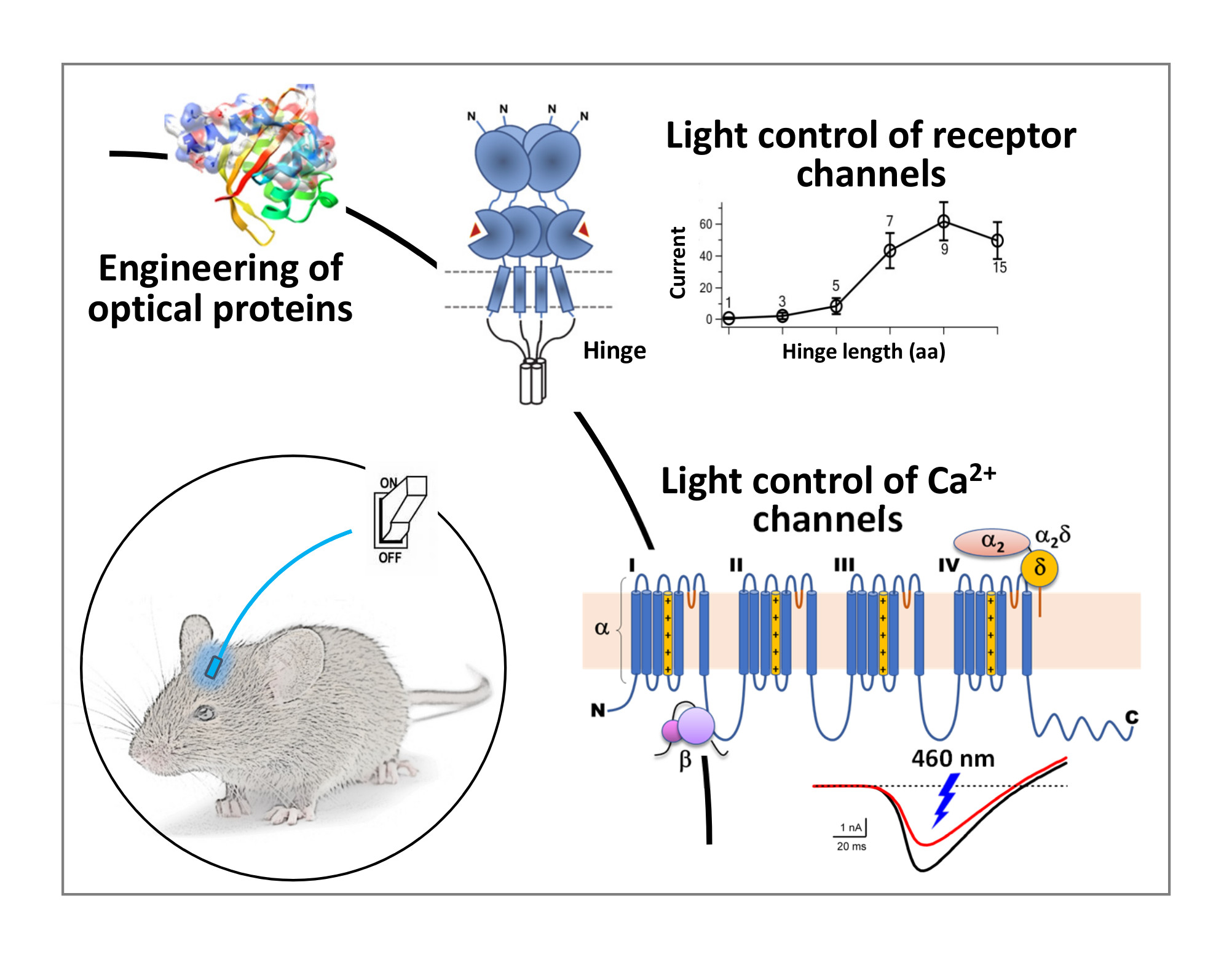
Optogenetic Control of Endogenous Channels
Optogenetics uses light-gated ion channels to activate or inactivate neural activity and is a new frontier of neuroscience. The widely used microbial opsins are effective optogenetic tools for basic research but face uncertainty in medical applications due to their microbial origin and distinct physiology compared to endogenous ion channels. We have developed a structural framework to exert light control of receptor channels and re-engineered protein photosensors to regulate the function of voltage-gated Ca2+ channels with light. These light-controlled endogenous ion channels, when used with induced pluripotent stem cells (iPSCs), will have important implications for regenerative medicine.